I combine innovative ideas, large genomic data sets, and bioinformatics tools to create novel, interdisciplinary research programs.
Microbial tools for forensic Science
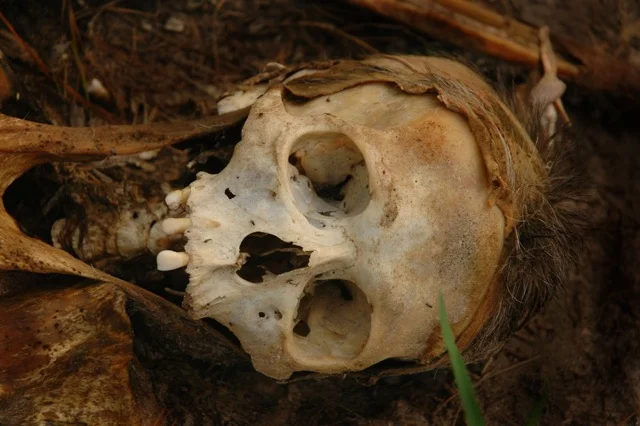
Microbial stopwatch of death
Because microbes are ubiquitous, they can be tiny witnesses to the events of our lives. I have been developing a microbial stopwatch that begins at death as a novel tool for estimating the time since death, or postmortem interval (PMI). PMI is important for criminal investigations because it can lead to the identification of the deceased and validate alibis. However, establishing PMI can be very challenging with few evidentiary tools available after the first after death. Rob Knight (UCSD), David Carter (Chaminade University), and I, along with a number of excellent collaborators, received funding from the National Institutes of Justice (NIJ 2011-DN-BX-K533, 2014-R2-CX-K01, 2015-DN-BX-K016) to support research to test and fine tune the use of microbiome tools for estimating PMI. Please read about it in our most recent publication in the journal Science.
Solving Crimes with the Necrobiome
My lab was recently featured in a segment narrated by Ed Yong, the acclaimed science writer who authored "I Contain Multitudes", which lets us peek into the ubiquitous nature of microbes and how vital they are. Check it out below!
The ancestral gut microbiome
What is a healthy human gut microbiome? We are currently characterizing the human gut microbiome in cultures that have dramatically altered their microbial counterparts with intensive hygiene practices, widespread antibiotic use, high fat and protein diets, and extensive time spent indoors. We can try to understand the ancestral state of the human gut microbiome in two ways to do this – 1) study the microbiome of human populations living traditional lifestyles, particularly ones that haven’t been exposed to antibiotics and 2) by sampling ancient gut microbiomes in well preserved ancient human fecal material. I currently work on both of these types of projects. Through highly collaborative research projects, I study the gut microbiomes of people living traditional lifestyles today (e.g. hunter-gatherers). Additionally, I study gut microbiota preserved in ancient fecal material discovered in caves, Viking latrines, and naturally mummified human remains.
The effect of domestication and captivity on the microbiome
The Przewalski horse microbiome
We are only just beginning to understand the complex and beautiful relationship between vertebrate hosts and their microbial partners. I am interested in a broad range of questions about how vertebrate microbiomes have evolved and how they may have changed through domestication and modern agriculture. Recently I worked with an international group of collaborators to study the difference between domesticated horses and Przewalski horses, which are the only group of horses left on earth not successfully domesticated by humans. I had the opportunity to work with Claudia Feh (Association pour le cheval de Przewalski: TAKH, Station biologique de la Tour du Valat) and Ludovic Orlando (Laboratoire d’Anthropobiologie Moléculaire et d’Imagerie de Synthèse, Université de Toulouse). Their teams collected fecal samples and important metadata from herded, domestic horses and Przewalski horses living near Seer, Mongolia and separated by a fence.
Check out our recent paper published in Scientific Reports.
We discovered Przewalski horse fecal microbiomes hosted a distinct and more diverse community of bacteria compared to domestic horses, which is likely partly explained by different plant diets as revealed by trnL maker data. Within the PH population, four individuals were born in captivity in European zoos and hosted a strikingly low diversity of fecal microbiota compared to individuals born in natural reserves in France and Mongolia. These results suggest that anthropogenic forces can dramatically reshape equid gastrointestinal microbiomes, which has broader implications for the conservation management of endangered mammals.
Here we link to an updated Supplementary Table 3, in which we added and corrected several items highlighted in red. You can find a description of the changes in the comment section of our online manuscript.
photo by Ludovic Orlando
rumen microbiomes
I am also very interested in rumen microbiomes and how rumen ecology of domesticated livestock may differ from their wild ancestors or close relatives. I am starting several projects along these lines.
Tumor microbiomes
LONGITUDINAL STUDY OF THE MICROBIOME OF APPENDICEAL CANCER PATIENTS
Appendix cancer is newly diagnosed in over 1,000 Americans a year. Recent research has revealed the presence, and potential role, of bacteria in tumor material of the appendiceal neoplasm Pseudomyxoma Peritonei (PMP) (1), which is a mucinous tumor type invades the peritoneal cavity. Gilbreath et al. (2013) showed that tumor/mucin microbiome was dominated by bacteria of the Phylum Proteobacteria such as Helicobacter. Helicobacter pylori has been implicated in other gastric cancers with antibiotic treatments effectively reducing tumor and improving survivorship in some cases. Similarly, a pilot study completed by the authors of this proposal demonstrated that antibiotic treatment increased survivorship for patients with the malignant form of PMP (Gilbreath et al. 2013, Semino-Mora et al. 2013), which is also associated with higher bacterial loads.
We hypothesize that tumor-promoting species are more prevalent in the fecal flora of PMP patients at the time of diagnosis, and these species could serve as both diagnostic and therapeutic targets. We will generate longitudinal gut microbiome data from the first 25 patients enrolled in clinical trial NCT02387203 and determine whether microbiome composition influences either carcinogenesis or post-surgical diarrheal illness, with the potential of markedly improving the quality of life of patients.
My Postdoc and PhD research
Extinctions of South American megafauna
One of my passions is the study of animals that became extinct during the late Pleistocene. What are their closest extant relatives? When did they became extinct? Were populations declining before extinction or did they became extinct suddenly? Is the timing of their extinction or population decline associated with changes in climate or human occupation of the region? A combination of radiocarbon data and DNA sequence data derived from teeth, bones, and other biological material provides clues to the answers of these questions. In collaboration with archaeologists and paleontologists, these data can be interpreted to help answer long-standing questions about the intriguing megafauna that no longer roam the earth. During my post-doc at ACAD, I worked on several projects in which we examined the late Pleistocene genetic diversity of large herbivores across a number of continents. In my focal study system of South American camelids, I uncovered previously undocumented extinction events in the Patagonia region of South America.
Conservation genetics of Colorado's greenback cutthroat trout
We clarified the taxonomy and diversity of cutthroat trout subspecies in the drainages of Colorado by using early museum collections (1857 – 1890 AD) of cutthroat trout. In my PhD research, I studied the population genetics and phylgenetics of modern cutthroat trout populations in Colorado, and demonstrated that early 20th century fish propagation and stocking activities had resulted in divergent lineages of cutthroat trout mixing across major drainage systems on both the Atlantic and Pacific slopes of the Continental Divide. The results demonstrated uncertainty about whether populations of the federally protected greenback cutthroat trout still existed (Metcalf et al. 2007). Subsequently, with funding and support from a wide range of federal and state agencies as well as Trout Unlimited, I successfully sequenced DNA recovered from 30 individuals of ~150 year old cutthroat trout specimens preserved in ethanol. The results of this research revealed recent extinctions, undescribed lineages, errors in taxonomy, and dramatic range changes induced by human movement of fish (Metcalf et al. 2012). We discovered that cutthroat trout lineages native to the Arkansas River and San Juan River, respectively, had become extinct since historic times. Our biggest surprise was the discovery of a single remaining population of greenback cutthroat trout (O. c. stomias) currently located outside of its native range in Bear Creek near Pikes Peak in the Arkansas River drainage. On August 8, 2014, in an effort spearheaded by the greenback cutthroat recovery team, our state fish was reintroduced to its native range.